LigAlign
Lilien Lab
Department of Computer Science
Centre for Cellular and Biomolecular Research
University of Toronto
Automated Ligand-Based Active Site Alignment
Proteins which bind the same ligand are likely to possess a conserved local protein-ligand interface even though they may not exhibit a conserved global architecture. We utilize bound ligand structures to compare the active sites of several proteins which interact with a chosen ligand. Our tool automates a constrained version of structural alignment: beginning with a ligand, it gathers a set of experimental structures of proteins bound to the ligand from the Protein Data Bank. The tool then aligns the ligands bound in the structures according to minimum RMSD. Finally, the user can visually examine the aligned active sites to identify structural patterns, such as conserved steric hindrance or hydrophobicity. Similarly, portions of the active site which demonstrate high variation across proteins may not strongly affect binding to the ligand.
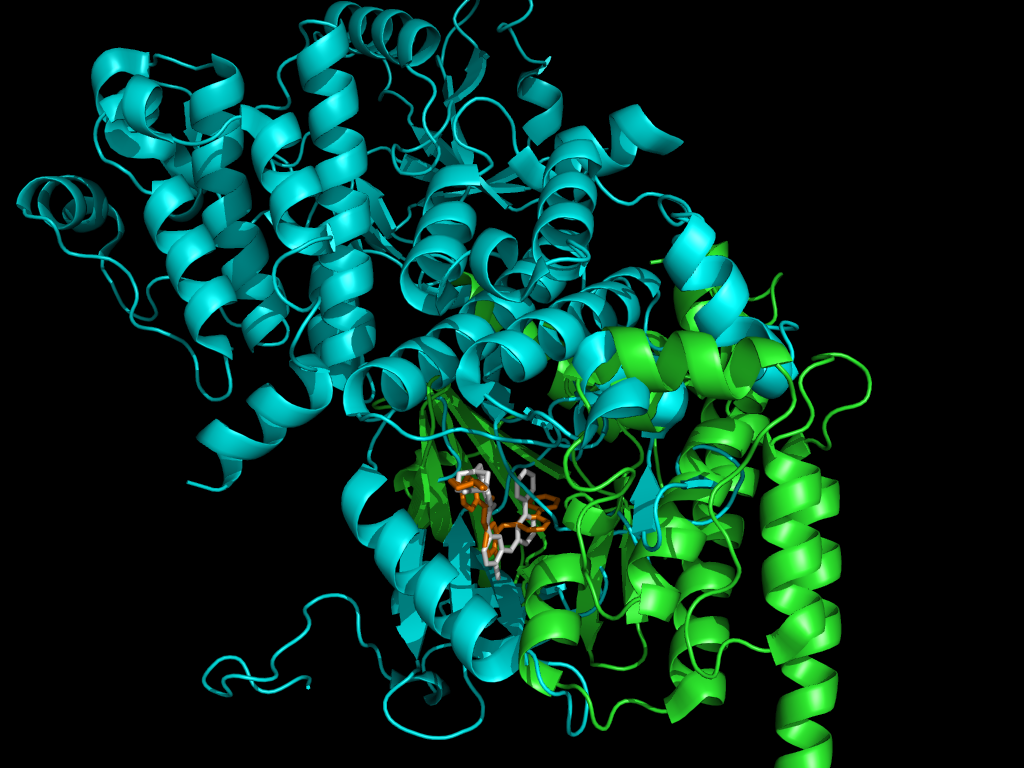
Protein Alignment versus Ligand Alignment
Although proteins which bind the same ligand may not exhibit a conserved global architecture, they are likely to possess a conserved local protein-ligand interface. We utilize bound ligand structures to compare the active sites of proteins which interact with a chosen ligand. The figure above shows the Gleevec-based alignment of two proteins.
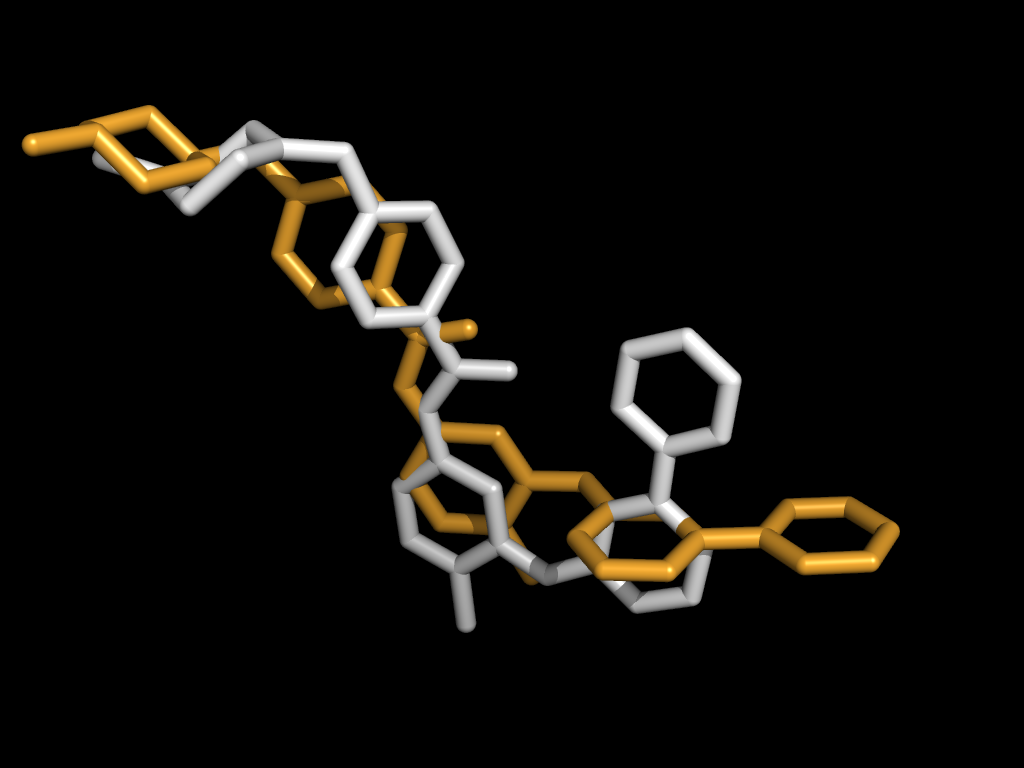
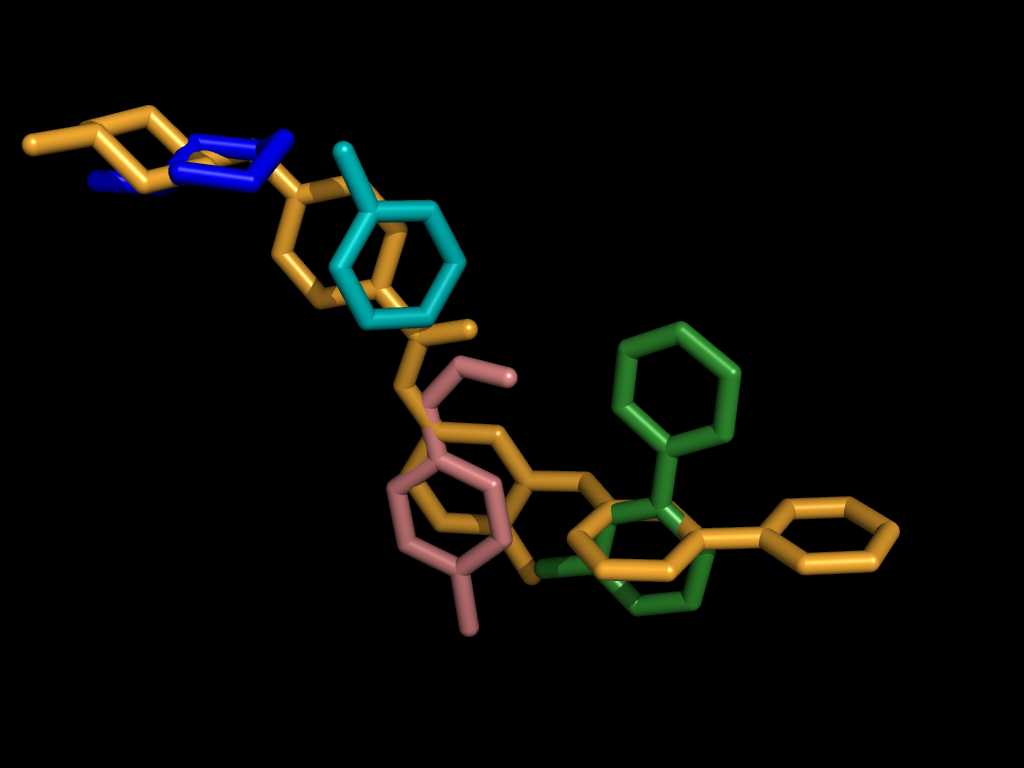
| 
|
Ligand Flexibility
Given the structures of multiple protein-ligand complexes, we can align the active sites by properly aligning the ligands. The simplest method to align two ligands is a rigid transform but flexible molecules dock in a variety of conformations. This variability hinders comparison of active sites.
We can enforce a correspondence across conformations by aligning ligand fragments. Given two ligands, one acting as a pivot (above left, colored white) and one query to be fragmented (above left, orange), we can automatically break the query into fragments (above right) and independently align each.
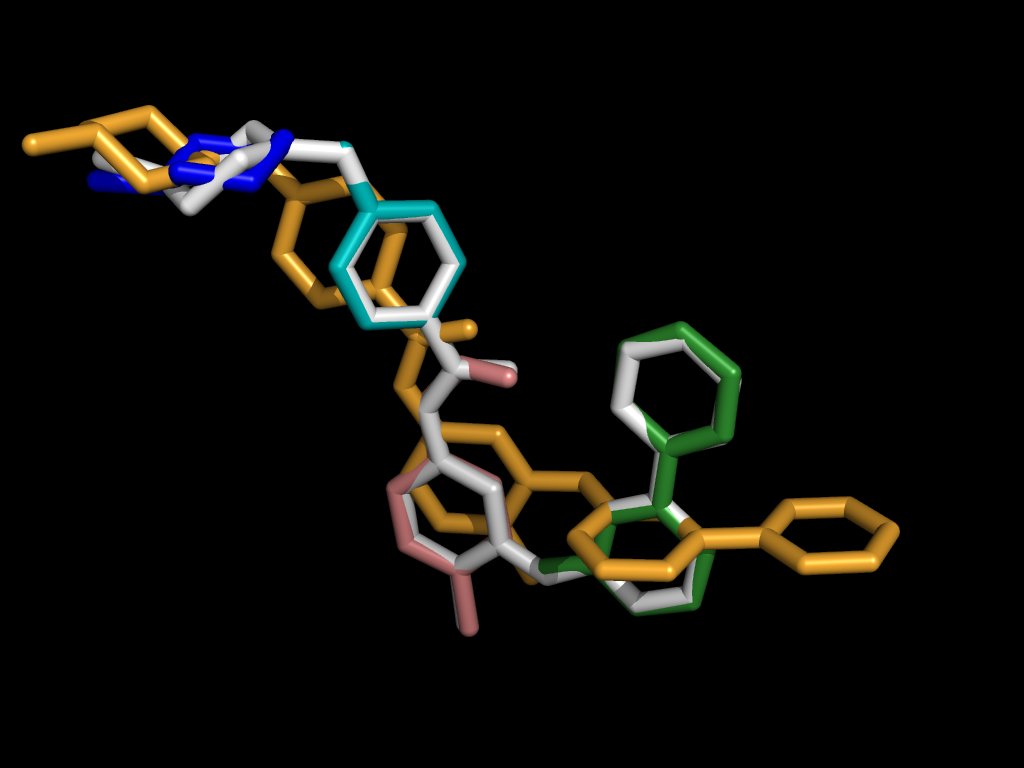
A Fragment-based Approach
As we increase the number of fragments, the difference between the fragmented query and the pivot decreases.
For each fragment, this generates an active site alignment based on the coordinate frame of only that fragment. This simplifies the comparison of local features within the active sites of multiple proteins.
LigAlign was first developed by Abraham Heifets and Ryan Lilien.