LigAlign
Lilien Lab
Department of Computer Science
Centre for Cellular and Biomolecular Research
University of Toronto
Documentation
Example scripts
If you want to get aligning as quickly as possible, download and run the following scripts to reproduce our heme and NAD experiments:Tutorial
This tutorial demonstrates LigAlign's rigid and flexible alignments of Gleevec (imatinib) binding proteins.-
Load LigAlign. Begin by running PyMOL on the ligand_alignment.py extension. For example, start PyMOL, at the PyMOL prompt (in the PyMOL program), change into the directory containing the ligand_alignment.py file (to do this, type "cd directoryname" without the quotes and with your directory name in place of 'directoryname'). When you are in the correct directory, type the following command into the PyMOL prompt,
run ligand_alignment.pyThe text "LigAlign v1.00 loaded." should appear. If this welcome message does not appear, you are most likely not in the correct directory - seek the friendly help of your local computer guru. -
At the PyMOL prompt, we can begin by running:
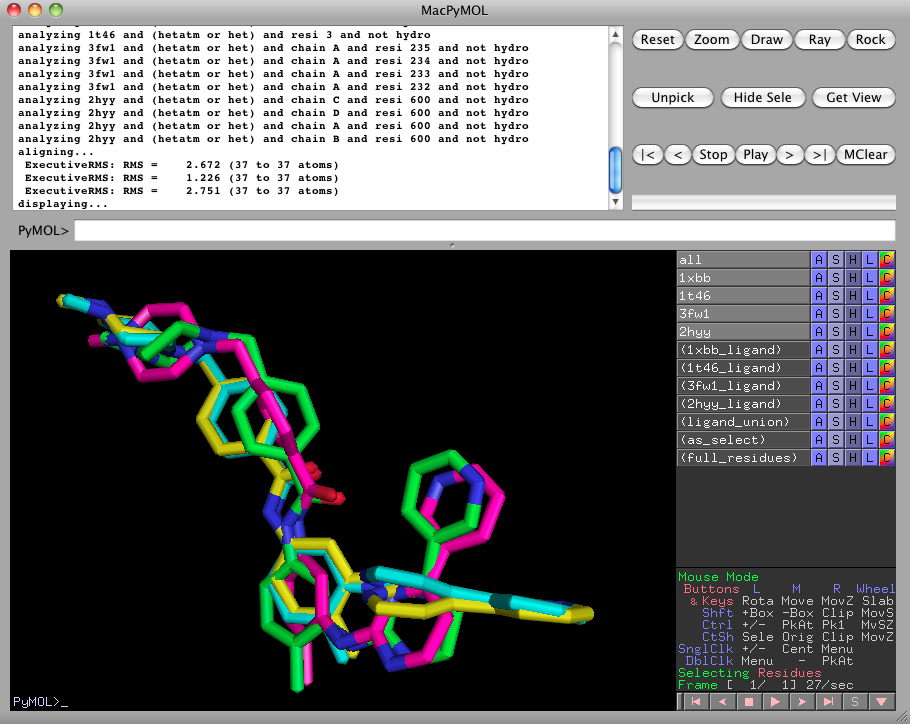
ligalign 1xbb, 1t46, 3fw1, 2hyywhich will align 1t46, 3fw1, and 2hyy against 1xbb so as to minimize the RMSD between the bound imatinib ligands. This may take a few moments to fetch the PDB files.
-
As we can see, LigAlign populates the right pane with a number of interesting selections. For now, hide the 3fw1 and 2hyy proteins by clicking on 3fw1 and 2hyy in the right pane.
Then run
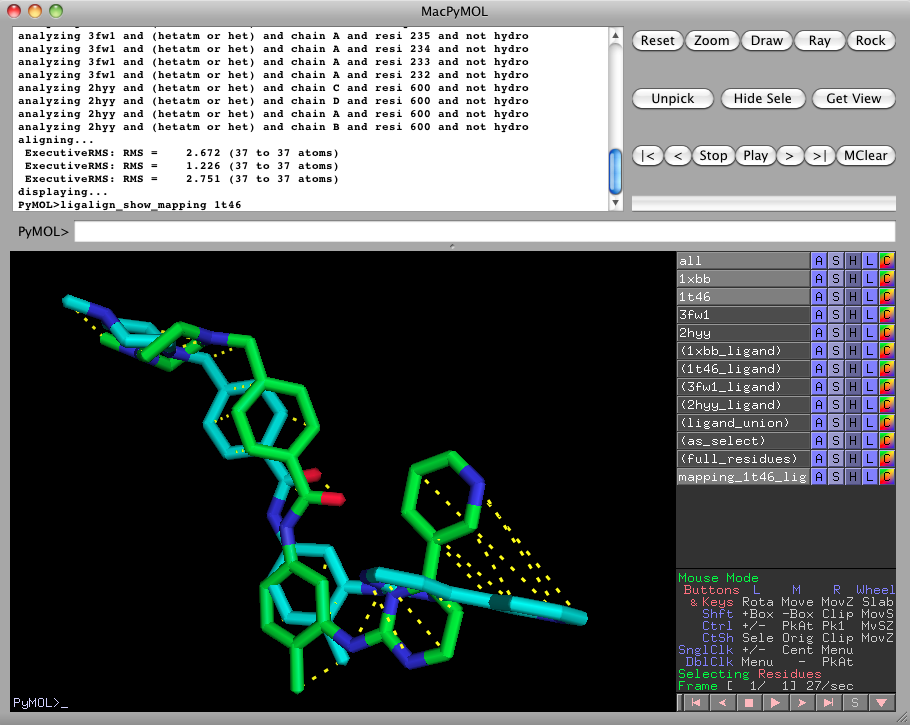
ligalign_show_mapping 1t46to show the atom-to-atom mappings computed by LigAlign. All of the atoms of 1t46 are shown mapped to the corresponding atoms within 1xbb (because it was the pivot-selection from the last executed ligalign command (in step 1)). When we are done viewing the atom mappings, turn off the atom mapping by clicking on mapping_1t46_ligand_to_1xbb_ligand in the right pane.
-
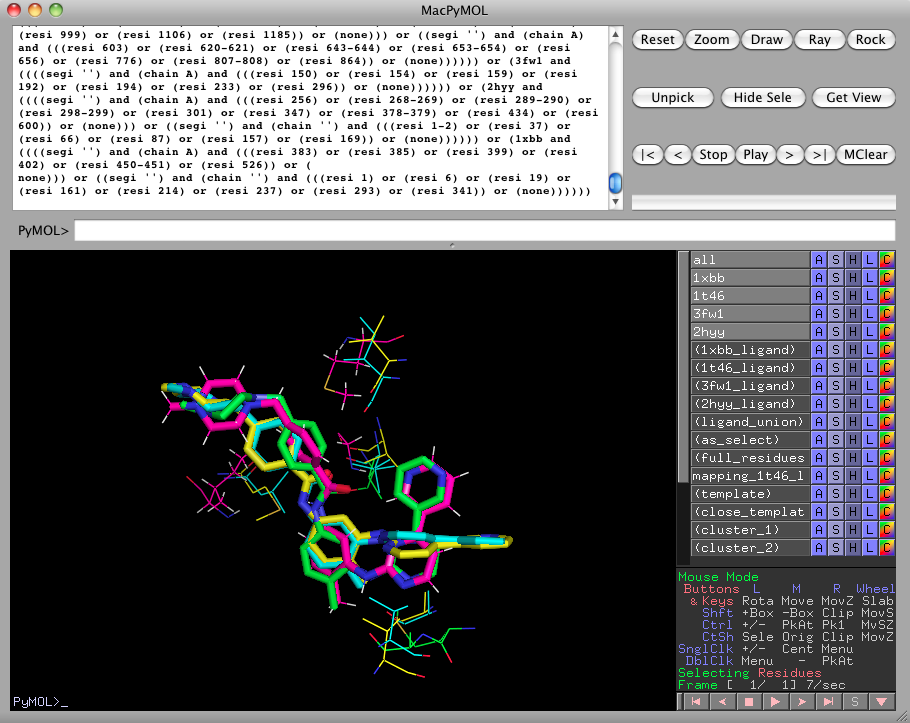
We'll now look for consistent clusters of amino acids in the protein active sites. Begin by showing 3fw1 and 2hyy again, by clicking on 3fw1 and 2hyy in the right pane. Now run
ligalign_find_template 3.0, 0.75, chemical, 1xbb, 1t46, 3fw1, 2hyyThis looks for clusters with a diameter of 3.0 angstroms, consisting of residues from at least 3 out of 4 (i.e., 75%) of the listed proteins. All of the residues in a cluster must be in the same chemical class. By default, ligalign_find_template depicts the clusters within 4.5 angstroms as lines. If you'd like to see the clusters detected further away from the ligands, use the "template" selection or change the default "close_template" selection distance with ligalign_set.
-
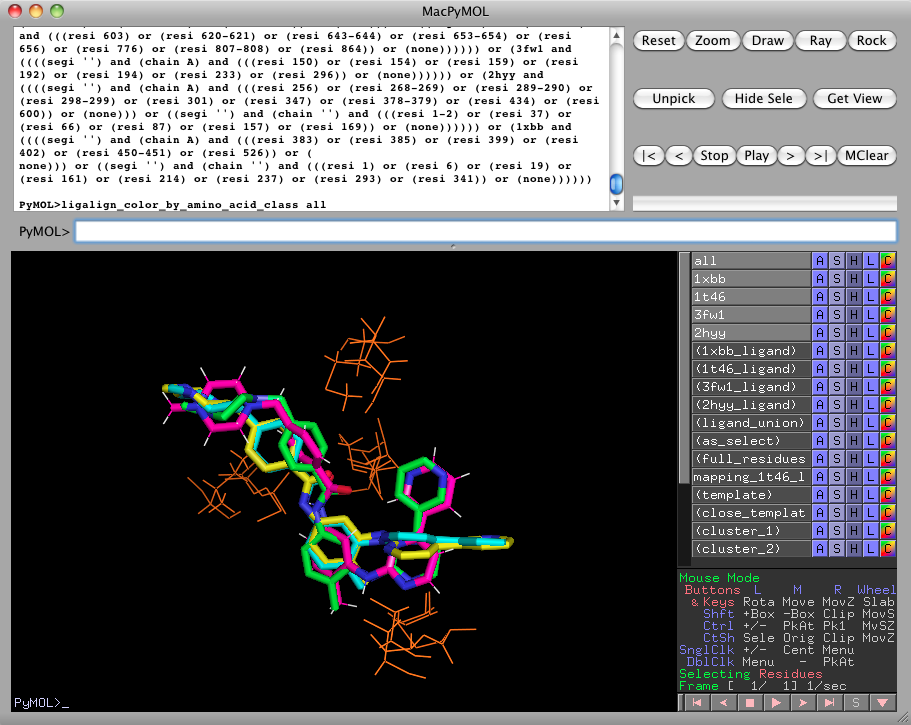
It can be a little difficult to detect patterns when the protein side-chains are inconsistently colored. Running
ligalign_color_by_amino_acid_class allwill color the side-chains according to their chemical class. Here, we see that the active site clusters are nonpolar. See the description of ligalign_color_by_amino_acid_class for chemical classes and colors.
-
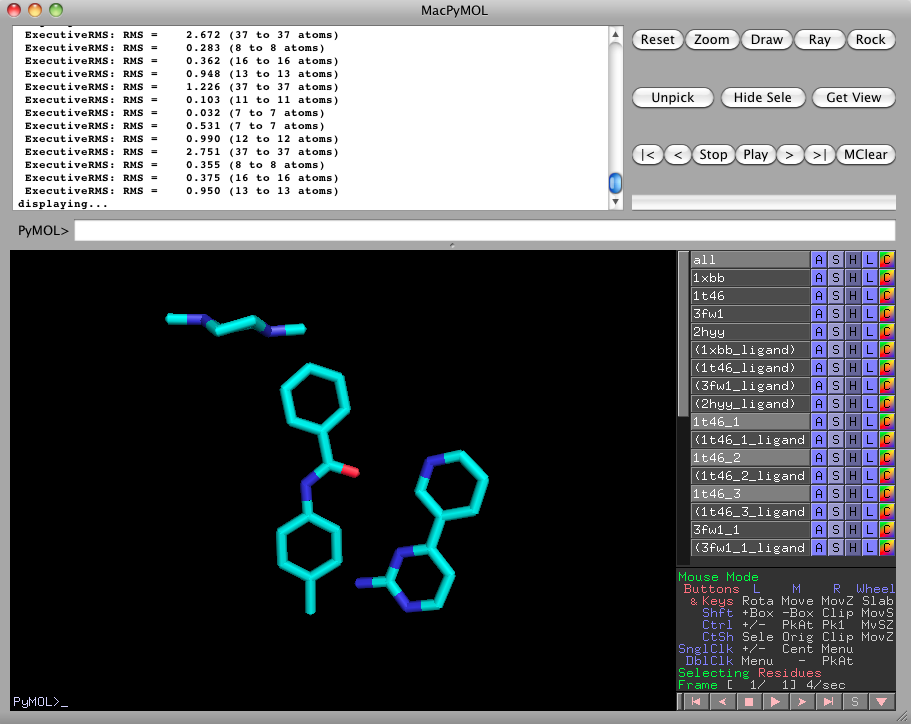
Clearly, the imatinib ligand has internal degrees of freedom. The ligands in 1xbb and 1t46 do not superimpose well. Perform a substructure-based alignment with
ligalign_fragments 1xbb, 1t46, 3fw1, 2hyyTurn off everything except for 1t46_1, 1t46_2, and 1t46_3. This is the fragmentation of the 1t46 imatinib ligand into three pieces.
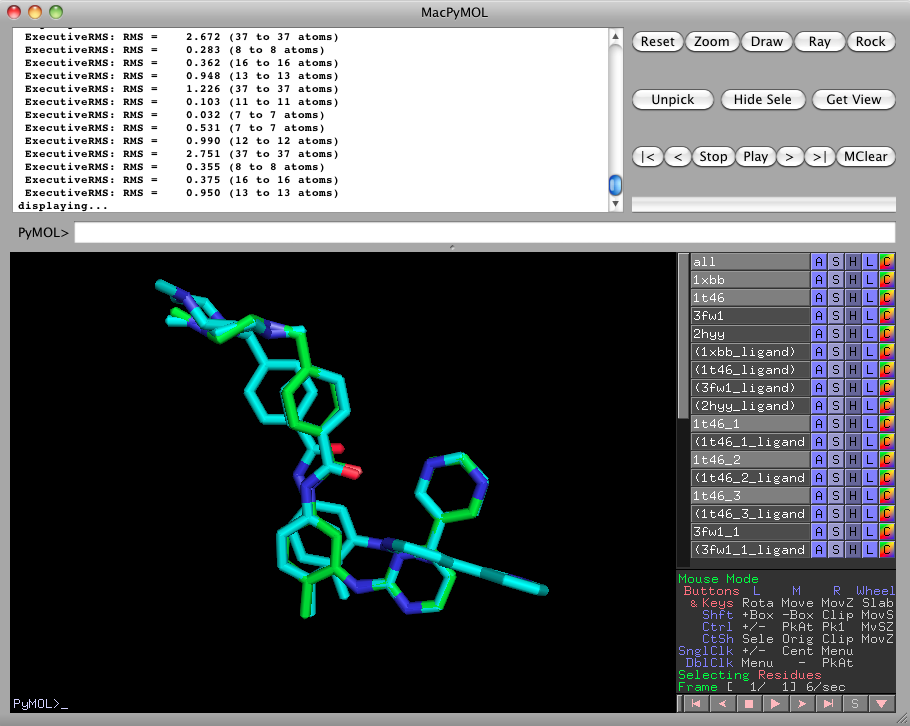
Please turn 1xbb and 1t46 back on, by clicking on them in the right pane. Notice how the fragments 1t46_1, 1t46_2, and 1t46_3 are aligned more closely to 1xbb than the rigid complete 1t46 could be. -
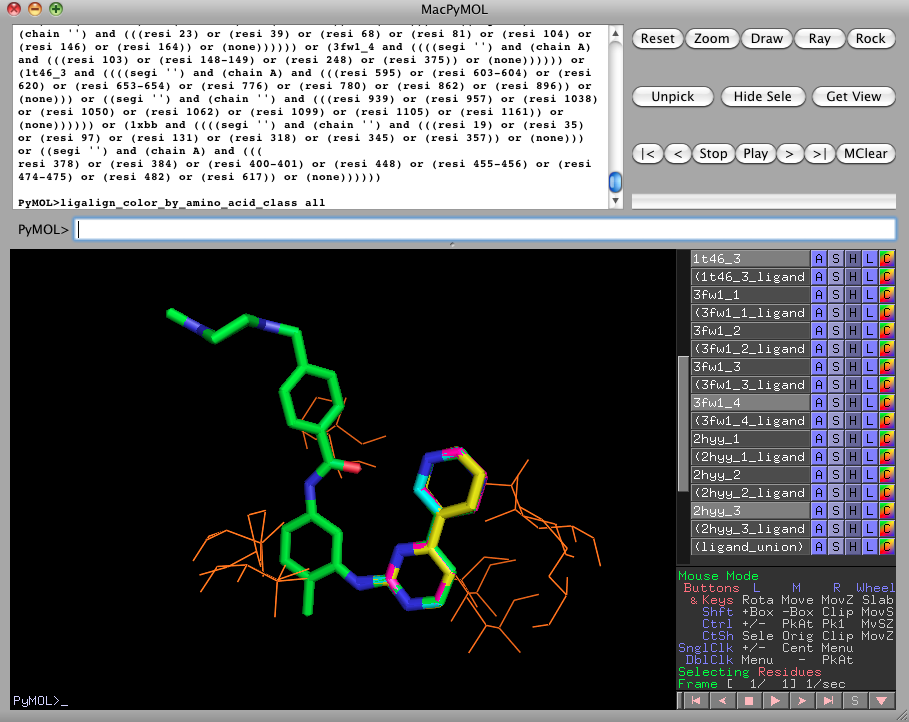
Now turn off every object except for 1xbb, 1t46_3, 3fw1_4, and 2hyy_3. These are the fragments containing the pyridine and aminopyrimidine rings of the imatinib ligands. We can search for active site clusters based on this superposition with the following series of commands:
ligalign_find_template 3.0, 0.75, chemical, 1xbb, 1t46_3, 3fw1_4, 2hyy_3As expected, this produces a different set of clusters, as compared to the rigid alignment, because the new orientations of the bound proteins cause different subcavities of the active sites to superimpose.
ligalign_color_by_amino_acid_class all
-
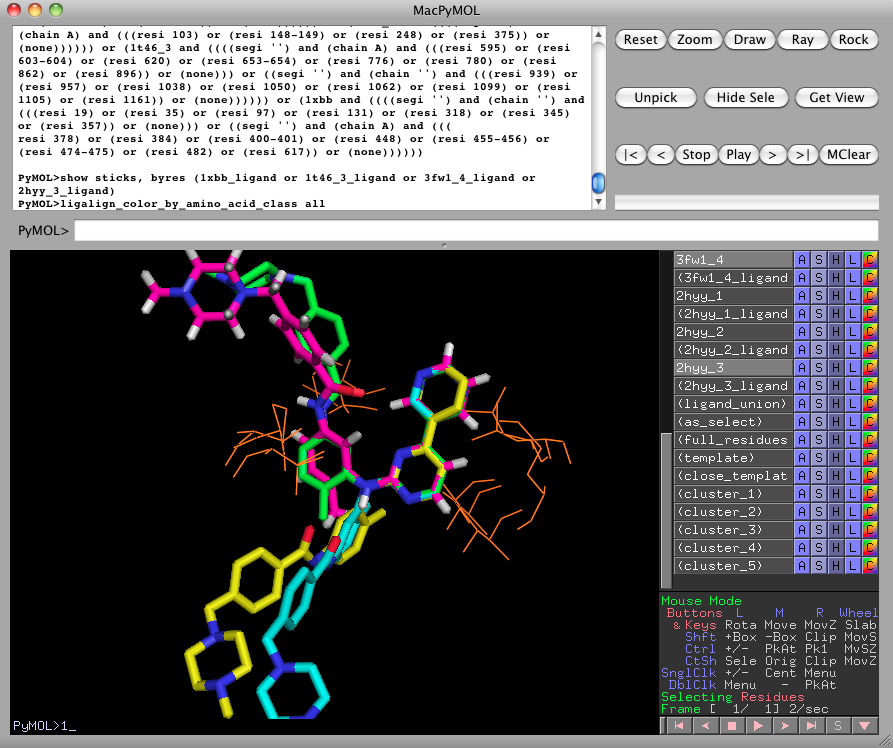
We can also display the full imatinib ligands with the following command:
show sticks, byres (1xbb_ligand or 1t46_3_ligand or 3fw1_4_ligand or 2hyy_3_ligand)This view highlights the disorder induced on distant ligand moieties, such as the N-methylpiperazine ring, when we focus on optimizing the alignment of one fragment.